9 Chapter 9 – Isolation of DNA
Isolation of DNA
BACKGROUND
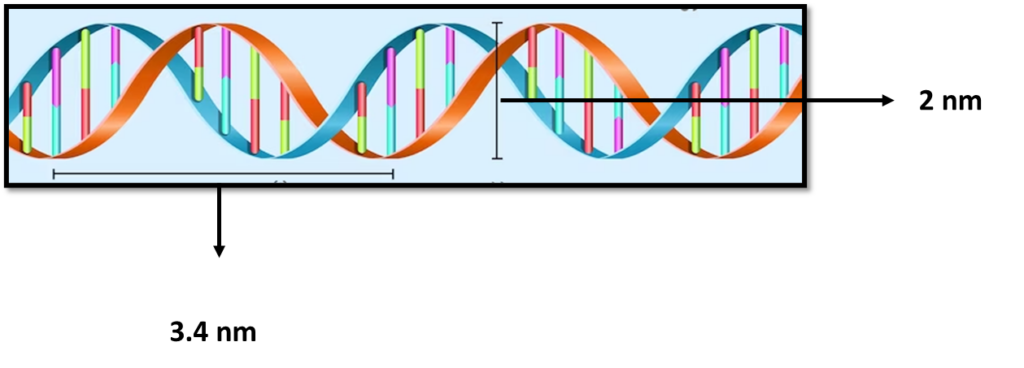
is a polymer composed of two polynucleotide chains that coil around each other to form a double helix structure. This polymer carries genetic instructions for all known organisms and many viruses.
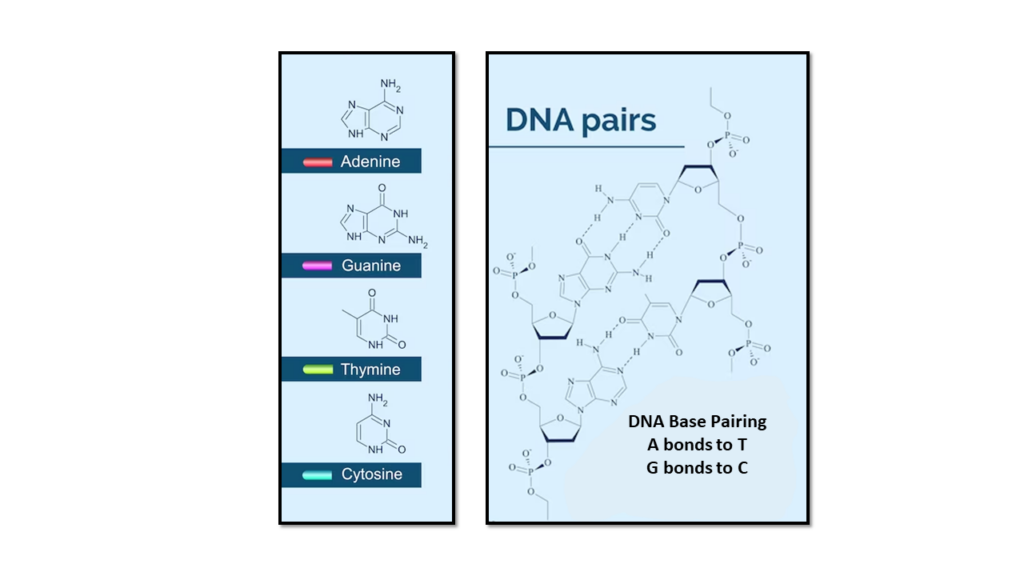
The two DNA strands are known as , composed of simpler monomer units called nucleotides. Each nucleotide is composed of one of four nucleobases (, , , or , a sugar called deoxyribose, and a phosphate group. The nucleotides are joined in a chain by phosphodiester linkage (covalent bonds) between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are held together with hydrogen bonds (A with T and C with G) to make double-stranded DNA.
The complementary nitrogenous bases are divided into two groups, pyrimidines, and purines. In DNA, the pyrimidines are and ; the purines are and .
A large part of (more than 98% for humans) is non-coding, meaning these sections do not serve as patterns for protein sequences. The two strands of DNA run in opposite directions and are thus antiparallel. Both the strands of double-stranded DNA store the same biological information.
DNA isolation and purification are used in laboratories engaged in molecular biology experiments. Several standardized techniques and variations are adapted according to the type of cells or tissues. The process involved lengthy and tiresome ultra-centrifugation in the early days of DNA isolation and purification methods. Now, with the advancement of separation techniques, the procedure is short and agile.
In any method of extraction and purification, there are three main steps: breaking the cells, DNA extraction, and purification.
Cells are broken in different ways depending on the cell type. One standard method for lysis of bacterial cultures is alkaline lysis. In the case of animal cells, lysis is accomplished by detergents or hypotonic solutions.
Plant tissues are homogenized by strong detergents such as SDS (sodium dodecyl sulfate) and heated at high temperatures. Various DNA isolation kits are sold by several biotechnology companies, which are very simple, short, and easy to handle.
The isolation of bacterial plasmid DNA by alkaline lysis method is used for the large-scale isolation of plasmid DNA by modification of the alkaline lysis procedure, followed by purification by phenol-chloroform extraction. Cells containing the desired plasmids are harvested by centrifugation, incubated in lysozyme buffer (re-suspension buffer), and treated with an alkaline detergent. The alkali breaks the cells, releasing DNA and proteins into the medium. Detergent solubilizes the proteins and DNA. The proteins and membranes are precipitated with sodium acetate. The precipitate is centrifuged at a higher RPM, and the supernatant contains the DNA. Finally, the DNA is precipitated by adding 95% ethyl alcohol or propanol. The DNA pellet is re-suspended in a Tris-EDTA (TE) buffer. This DNA sample contains some DNA-binding proteins, which have to be removed. This procedure is often done by phenol-chloroform extraction. This protocol has several variations, each suited to the situation and type of bacterial culture.
Genomic DNA isolation is performed according to the standard protocol suggested by the Federal Bureau of Investigation (USA). The blood samples are stored at -70°C in EDTA vacutainer tubes. Once thawed, a standard citrate buffer is added, the tubes are mixed, then centrifuged. The top portion of the supernatant is discarded, and the additional buffer is added. The tubes are again mixed and centrifuged. The supernatant is discarded, and the pellet is re-suspended in a solution of SDS detergent and proteinase K. This mixture is then incubated at 55°C for one hour. Then the sample is phenol-extracted once with phenol/chloroform/isoamyl alcohol solution and centrifuged. The aqueous layer is removed to a fresh microcentrifuge tube. The DNA is ethanol-precipitated, re-suspended in buffer, and ethanol-precipitated a second time. After the pellet is dried, the buffer is added, and the DNA is re-suspended by incubation at 55°C overnight. A polymerase chain reaction later assays the genomic DNA solution.
Plant tissues bring up several problems during DNA isolation. Plant cells have a rigid cell wall, and the tissue contains many toxic metabolites that can interact with the DNA and change its nature, making it useless for other experimental purposes. Metabolites such as mucilage and other carbohydrates can quickly form complexes with DNA and can damage it. Therefore, the extraction buffer should be supplemented with compounds protecting DNA against these metabolites. Plant molecular biologists widely employ Many DNA isolation techniques using CTAB (Cetyltrimethylammonium Bromide) extraction buffer. This compound forms a complex with DNA and thus protects it from other toxic metabolites such as mucilage and phenolic compounds.
The DNA, isolated and purified by these methods, can be used for various experimental purposes. It can be used for restriction digestion analysis, cloning, ligation, transformation experiments, in vitro transcription, PCR amplification, RFLP (restriction fragment length polymorphism), fingerprinting, RAPID (random amplification polymorphic DNA), sequencing, nick translation and radio labeling, preparation of genomic DNA libraries and cDNA libraries, etc.
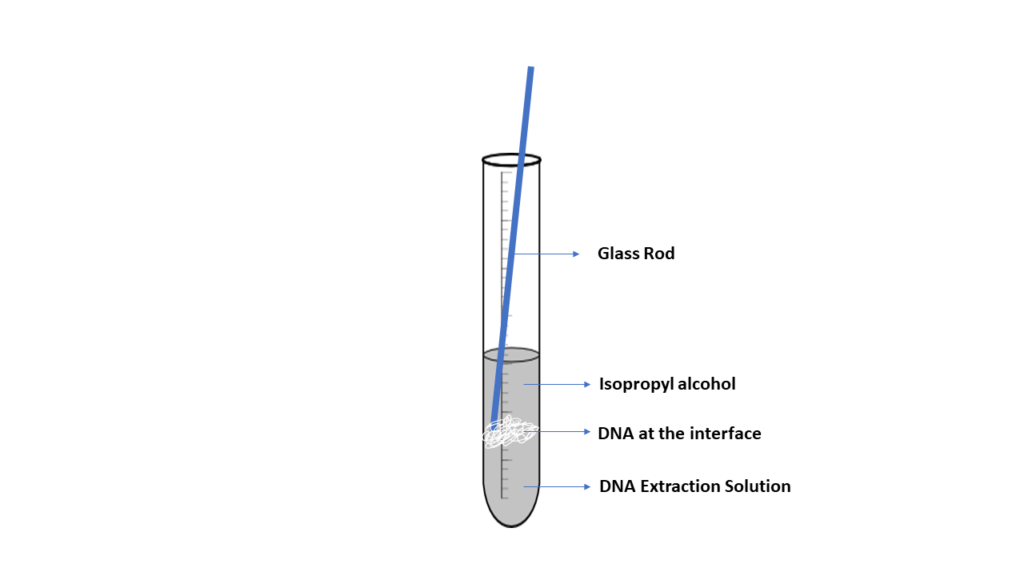
DNA is spooled together using alcohol, which allows DNA fragments to stick together, producing a blob of DNA. When a small layer of alcohol is added to a solution containing cellular fragments and DNA, it will form an interface where the DNA will precipitate. The DNA can then be captured or spooled onto a wooden stick or glass rod. Although this method is effective, the DNA produced is not pure. Other materials, such as protein and cell fragments, are present in the DNA.
Optional Review:
- Go to the Howard Hughes Medical Institute BioInteractive page
- Explore the history of DNA discovery.
- Explore DNA structure and how it relates to the function.
- Explore Central Dogma and Genetic Medicine with the interactive lab at HHMI
Key Terms
- Alkaline lysis method
- Lysis
- Supernatant
- Metabolite
- Extraction Buffer
- Supernatant
- Plasmid
- Precipitation
- Plasmid DNA
Objectives
- Extract DNA from cells and observe DNA molecules.
- Separate DNA from cell membranes and proteins by centrifugation.
- Precipitate dissolved DNA and spool the molecule out of solution.
- Observe DNA under a microscope (Optional).
Materials
- DNA extraction buffer*
- Diced fruit or vegetable**
- Mortar & pestle or blender
- Cheesecloth
- Wooden applicator or glass rod
- Centrifuge
- 95% isopropyl alcohol, on ice
- Test tube (15ml) or Falcon capped tube (2)
- Graduated cylinder (10ml)
- Methylene blue
- Pipet pump and 10ml pipets
-
- *To make DNA extraction buffer: combine 2.5ml detergent, 0.75g NaCl (non-iodized salt), 2.5g NaHCO3 (baking soda), and 60ml distilled water. Chill in an ice bath.
- **The DNA found in strawberry cells can be extracted using common, everyday materials. Strawberries are soft and easy to pulverize. Strawberries have large genomes; they are octoploid, which means they have eight of each type of chromosome in each cell. Thus, strawberries are an exceptional fruit to use in DNA extraction labs and strawberries yield more DNA than any other fruit (i.e. banana, kiwi, etc.).
- Strawberries can be substituted with kiwi, bananas, tomatoes, or green onions.
Pre-Assessment
- Watson and Crick developed a model of DNA. State 4 things that you know about the DNA model.
- What kind of bond holds the two antiparallel strands of DNA nucleotides?
- If we think of the DNA double helix as a twisted ladder, what makes up the rungs or steps of the ladder?
- The Deoxyribose sugar has a carbon atom that is not part of the pentose ring. In a nucleotide (purine or pyrimidine), what is attached to this carbon?
- What are the three main steps in any method of DNA isolation?
- What method would you use for the large-scale isolation of plasmid DNA?
- In the DNA isolation techniques, DNA is vulnerable and can be destryoed by metabolites. What compound is added to the extraction buffer to protech the DNA?
Exercise 1: DNA Extraction
PROCEDURE
- *IMPORTANT* You will need to submit pictures showing the stages of DNA extraction and include them with your report, labeling the supernatant, interface, and the isolated DNA. You may include additional lab appropriate images at your discretion.
- Make a pulp with a small section of fruit or vegetable and grind the tissue with deionized water. Use a blender or mortar and pestle to grind the tissue.
- Transfer 2.5ml of the ground tissue into a test tube or Falcon capped tube. Add 6.5 ml of DNA extraction buffer.
- Grind the tissue in the tube with a glass rod or use a vortex to disperse the tissue and release the cellular contents.
- Leave the sample at room temperature for 10 minutes.
- Spin the sample in the tube for 10 minutes at 2500 rpm in a centrifuge. Alternatively, filter the contents of the tube into a clean tube using cheesecloth and a funnel.
- Transfer 5 ml of the upper solution (supernatant) into a clean test tube. The DNA is contained in this portion.
- Carefully add 10 ml of chilled 95% isopropanol to the DNA solution, allowing the alcohol to stream slowly and gently along the inside of the test tube. The alcohol should float on top since the DNA/buffer solution is denser than the alcohol. The boundary between the two is called the interface.
- Insert the wooden applicator or glass rod into the test tube and swirl at the interface of the two liquids. The DNA will spool around the rod, appearing as a viscous, clotted mass.
- After about one minute of spooling, slowly remove the rod from the tube.
- Add a few drops of methylene blue stain to the solution remaining in the tube. This dye will stain any remaining DNA that did not spool onto the rod.
- Discard any solid material into the trash and wash all glassware when you have finished your observations.
Exercise 2: Bacteria DNA Extraction (Optional)
The purification of genomic DNA from bacterial cultures provides the basis for downstream molecular analysis, and this process is often achieved using commercially available kits. The following is a step-by-step procedure adapted from
Reference: Wright MH, Adelskov J, Greene AC. Bacterial DNA Extraction Using Individual Enzymes and Phenol/Chloroform Separation. J Microbiol Biol Educ. 2017 Sep 1;18(2):18.2.48. doi: 10.1128/jmbe.v18i2.1348. PMID: 28861145; PMCID: PMC5577976.
Materials
| Reagent | Concentration | Description |
|---|---|---|
| RNase A (ThermoFisher Scientific) | 100 μg/mL | Degrades single stranded RNA. Buffer P1 is a resuspension buffer comprising 50 mM Tris-Cl (pH 8.0), 10 mM EDTA |
| Achromopeptidase (Sigma Aldrich) | 50 kU/mL | Lysis enzyme with strong bacteriolytic activity against gram-positive bacterial cell walls |
| Lysozyme (Sigma Aldrich) | 24,000 kU/mL | Lysis enzyme with bacteriolytic activity against gram-negative bacterial cell walls |
| Sodium dodecyl sulfate (SDS) | 10% | Solubilization of cell membrane lipids |
| Proteinase K (ThermoFisher Scientific) | 20 mg/mL | Digestion of proteins |
| Phenol:Chloroform:Isoamyl alcohol (PCI) solution | Separation of DNA from other cellular components. Comprised of Phenol:Chloroform:Isoamyl alcohol in a 25:24:1 ratio |
|
| Ethanol | 100% | Precipitates DNA from solution |
| Tris-EDTA (TE) Buffer | Buffer solution used to store purified DNA comprised of 10 mM Tris (pH 8.0), 1 mM EDTA |
Bacteria: culture in mid to late log phase.
PROCEDURE
- Transfer 10 mL of mid-to late-log-phase culture to a falcon tube and pellet the cells through centrifugation at 7,500 rpm for 10 minutes. Discard the supernatant.
- Resuspend pellet with 467 μL RNase A in Buffer P1 and transfer to a 1.5-mL microcentrifuge tube. Add 8 μL lysozyme and 5 μL achromopeptidase, gently mix and incubate at 37°C for 60 minutes.
- Add 30 μL 10% SDS (sodium dodecyl sulfate) and 3 μL proteinase K, gently invert and incubate at 50°C for 60 minutes.Add 525 μL PCI (Phenol:Chloroform:Isoamyl) solution and mix for 10 minutes by gentle inversion. Centrifuge at 12,000 rpm for 15 minutes.
- Extreme care and personal protective gear (gloves, lab coats, and safety goggles) should be used when working with phenol as it is corrosive and may cause severe burns. This step should be completed in a fume hood.
- Transfer the upper aqueous phase to a sterile 1.5-mL microcentrifuge tube, taking care not to disturb the bilayer.
- Add an equal volume of −20°C 100% ethanol and gently mix by inversion. Centrifuge at 12,000 rpm for 20 minutes.
- Carefully decant the supernatant and thoroughly dry pellet at room temperature or in a 50°C incubator.
- Over drying will result in making the DNA pellet more difficult to dissolve back into solution. The pellet may or may not be visible to the naked eye.
- Resuspend the pellet in 50 μL TE (Tris-EDTA) buffer and allow pellet to sit overnight at 4°C.
- Confirm presence and concentration of bacterial DNA by running 5 μL of product on a 1.5% agarose gel. Purified DNA will appear as a defined band when visualized under UV light.
DATA ANALYSIS & CRITICAL THINKING QUESTIONS
- Using your knowledge of the molecular components of a cell, explain the purpose of each component of the extraction buffer:
- salt
- detergent
- baking soda
- What is the purpose of grinding the tissue, either with a blender or with mortar and pestle?
- What does the cold ethanol do?
- Why can’t we use room temperature ethanol?
- What cellular components are removed in the pellet after the sample is centrifuged?
- Why can you not see the double helix when observing the DNA under a microscope?
- What is the function of protease?
- What does adding alcohol to the DNA solution accomplish?
- What physical property of DNA causes it to rise to the top of the filtrate?
- Briefly describe two tests that can be performed with the extracted DNA.
- Include appropriate images in your report. Be sure to label the supernatant, interface, and isolated DNA.
Licenses and Attributions
Biology I Cellular Processes Laboratory Manual by The authors & Hillsborough Community College is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The molecule inside cells that contains the genetic information responsible for the development and function of an organism. DNA molecules allow this information to be passed from one generation to the next. DNA is made up of a double-stranded helix held together by weak hydrogen bonds between purine-pyrimidine nucleotide base pairs: adenine (A) paired with thymine (T), and guanine (G) paired with cytosine (C). Also called deoxyribonucleic acid.
A polynucleotide is a combination of nucleotide monomers which are connected to each other through covalent bonds. A single polynucleotide molecule consists of 14 or more monomers of nucleotide in a chain structure
Cytosine is one of the four nucleobases found in DNA and RNA. It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached. The nucleoside of cytosine is cytidine. In base pairing, it forms three hydrogen bonds with guanine.
Guanine is one of the four nucleobases found in DNA and RNA. It is a purine nucleobase. The guanine nucleoside is called guanosine. In base pairing, it forms three hydrogen bonds with cytosine.
Adenine is one of the four nucleobases found in DNA and RNA. It is a purine nucleobase. In base pairing, it forms two hydrogen bonds with thiamine. Adenine is also found in the energy-rich adenosine triphosphate (ATP) and the cofactors nicotinamide adenine dinucleotide (NAD), flavin adenine dinucleotide (FAD) and Coenzyme A.
Thymine is one of the four nucleobases in the nucleic acid of DN. It is a pyrimidine nucleobase. Thymine is also known as 5-methyluracil,. In RNA, thymine is replaced by the nucleobase uracil. In base pairing, it forms two hydrogen bonds with adenine.
Escherichia coli (abbreviated as E. coli) are bacteria in people's and animals' environments, foods, and intestines. E. coli is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium.
If you work in a lab designated a BSL-1, the microbes such as E. coli do not pose health threats and present minimal potential hazards to laboratorians and the environment.
Although most strains of E. coli are harmless, others can make you sick. Some kinds of E. coli can cause diarrhea, while others cause urinary tract infections, respiratory illness, pneumonia, and other illnesses.

